House Price Prediction:
Part 1: Data Exploration
- Part 1: Data Exploration
- Objective:
- Import Python Packages:
- Import & Clean Data:
- PCA Analysis of the data:
- References:
I completed the WQU Machine Learning course 3 months ago and wanted to explore some new challenges. As a result I am exploring this Kaggle competition for leisure and am following a website cited in the references.
Objective:
Predict house prices
Import Python Packages:
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sb
import sklearn as sk
Import & Clean Data:
- Two data sets are provided one for testing and the other for training.
- We import each of the csv files into a pandas dataframe and remove any unwanted details
df_test = pd.read_csv('test.csv')
df_train = pd.read_csv('train.csv')
df_train.head()
| Id | MSSubClass | MSZoning | LotFrontage | LotArea | Street | Alley | LotShape | LandContour | Utilities | ... | PoolArea | PoolQC | Fence | MiscFeature | MiscVal | MoSold | YrSold | SaleType | SaleCondition | SalePrice | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 60 | RL | 65.0 | 8450 | Pave | NaN | Reg | Lvl | AllPub | ... | 0 | NaN | NaN | NaN | 0 | 2 | 2008 | WD | Normal | 208500 |
| 1 | 2 | 20 | RL | 80.0 | 9600 | Pave | NaN | Reg | Lvl | AllPub | ... | 0 | NaN | NaN | NaN | 0 | 5 | 2007 | WD | Normal | 181500 |
| 2 | 3 | 60 | RL | 68.0 | 11250 | Pave | NaN | IR1 | Lvl | AllPub | ... | 0 | NaN | NaN | NaN | 0 | 9 | 2008 | WD | Normal | 223500 |
| 3 | 4 | 70 | RL | 60.0 | 9550 | Pave | NaN | IR1 | Lvl | AllPub | ... | 0 | NaN | NaN | NaN | 0 | 2 | 2006 | WD | Abnorml | 140000 |
| 4 | 5 | 60 | RL | 84.0 | 14260 | Pave | NaN | IR1 | Lvl | AllPub | ... | 0 | NaN | NaN | NaN | 0 | 12 | 2008 | WD | Normal | 250000 |
5 rows × 81 columns
print(df_train.shape)
print(df_test.shape)
(1460, 81)
(1459, 80)
Visualise the data
- Apparently, the data has so many NaN data it may be wise not to drop them.
- Below we use a simple function to determine all the unique entries in the columns with many NaN values
- Since NaN is not a good key in a dictionary we will need to employ a work around for the possible NaN vales
- We notice that for the following columns the ‘NaN’ values are too many and we do not expect them to contribute significantly to the ML algorithm:
- Alley
- FirePlace
- PoolQG
- Fence
- MiscFeture
def unique_tally(data):
'''Returns a dictionary of the unique entries in a data column and their frequencies'''
isnan = list(data.isnull())
res = {}
for i in range(len(data)):
if isnan[i]:
key_ = 'NaN'
else:
key_ = data[i]
if key_ in res:
res[key_] +=1
else:
res[key_] = 1
return res
tallies = []
c = list(df_train.columns)
for col in c:
tallies.append(unique_tally(df_train[col]))
indx =[]
for k in range(len(tallies)):
if 'NaN' in tallies[k]:
indx.append(k)
len(indx)
plt.figure(figsize= (20,30))
for i in range(len(indx)):
plt.subplot(5,4,i+1)
plt.bar(range(len(tallies[indx[i]])), list(tallies[indx[i]].values()), align='center', label = c[indx[i]])
plt.xticks(range(len(tallies[indx[i]])), list(tallies[indx[i]].keys()), rotation=50)
plt.legend()
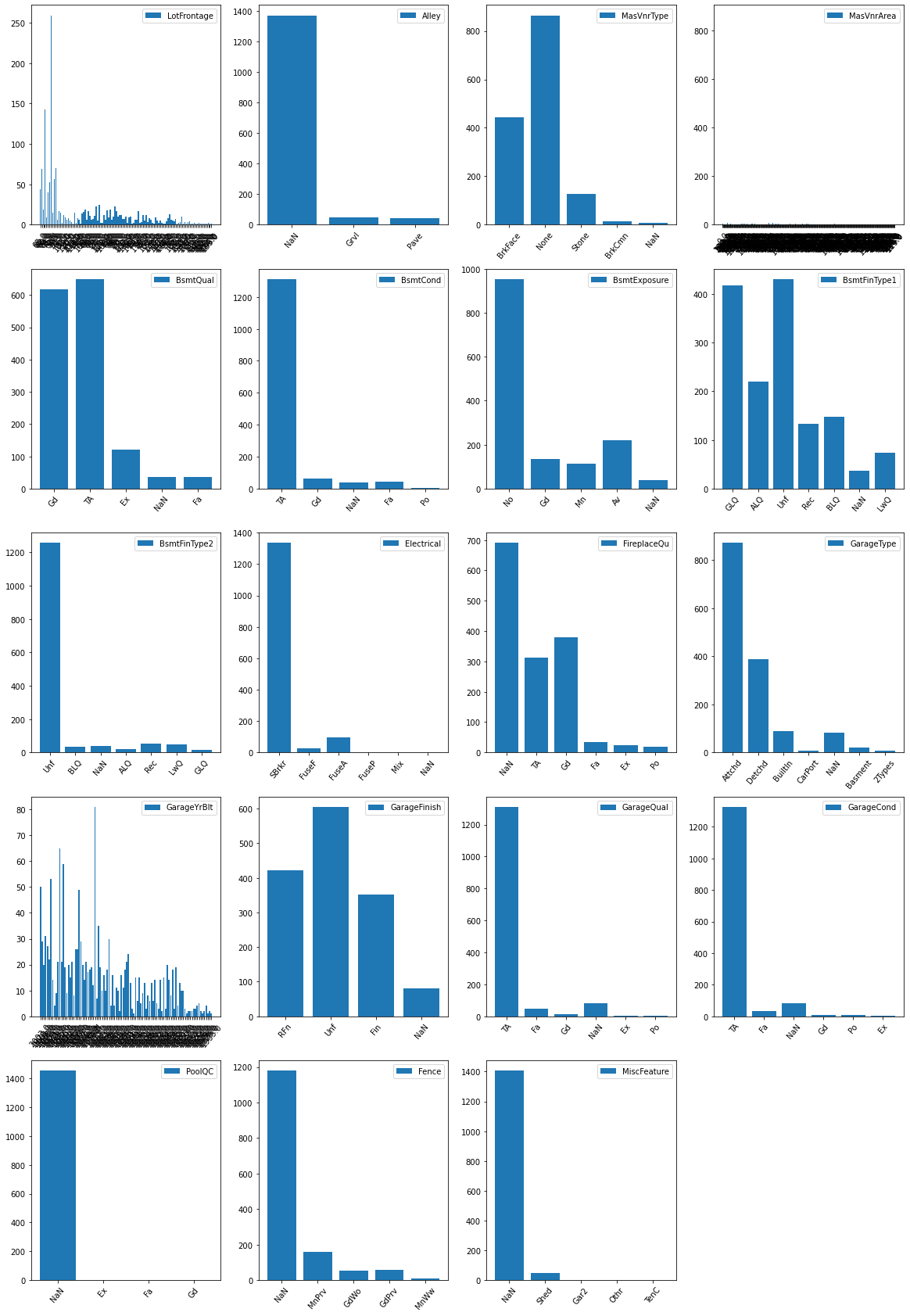
# Analyse the test data in a similar way using Pandas functions
df_train.isnull().sum().sort_values(ascending=False)
PoolQC 1453
MiscFeature 1406
Alley 1369
Fence 1179
FireplaceQu 690
...
CentralAir 0
SaleCondition 0
Heating 0
TotalBsmtSF 0
Id 0
Length: 81, dtype: int64
# Analyse the test data in a similar way using Pandas functions
df_test.isnull().sum().sort_values(ascending=False)
PoolQC 1456
MiscFeature 1408
Alley 1352
Fence 1169
FireplaceQu 730
...
Electrical 0
CentralAir 0
HeatingQC 0
Foundation 0
Id 0
Length: 80, dtype: int64
- So we will drop the following columns since they have more than 50% ‘NaN’ data values
- PoolQC
- MiscFeature
- Alley
- Fence
- Also, we will drop the ‘Id’ column as it is irrelevant to the calculations
df_train1 = df_train.drop(['Id', 'PoolQC', 'MiscFeature', 'Alley', 'Fence'], axis = 1)
df_test1 = df_test.drop(['Id', 'PoolQC', 'MiscFeature', 'Alley', 'Fence'], axis = 1)
Replacing NaN Values:
- Clearly, not all ‘NaN’ values need to be discarded.
- The data columns have various data types and we need to replace the these missing values in a consistent manner
- We do this for both the test and training data
- We will replace ‘NaN’ values depending on some conditions as follows:
- If the data in the column is numerical replace NaN with the mean
- If the data in the column is of string type, replace NaN with modal category
- We proceed as follows:
def replace_nan(df):
col = 0
c = list(df.columns)
for i in df.dtypes:
if i in [np.int64, np.float64]:
df[c[col]]=df[c[col]].fillna(df[c[col]].mean())
elif i == object:
df[c[col]]=df[c[col]].fillna(df[c[col]].mode()[0])
col+=1
replace_nan(df_train1)
replace_nan(df_test1)
sb.heatmap(df_train1.isnull(),yticklabels=False,cbar=False,cmap='coolwarm')

sb.heatmap(df_test1.isnull(),yticklabels=False,cbar=False,cmap='coolwarm')

Convert Categorical Data:
- All categorical data needs to be converted into numerical categories
- This will enable the algorithms to understand the data
def category_to_num(df):
'''Takes in a column of data and determines how many unique vakues there are
Each value is assigned a unique natural number & the data is updated
Returns the categories.'''
categs = sorted(list(df_train1[col[ci]].unique()))
for num in range(len(categs)):
df.loc[df==categs[num]] = num
return categs
ci = 0
col = list(df_train1.columns)
categ = {}
for dt in df_train1.dtypes:
if dt == object:
categs= category_to_num(df_train1[col[ci]])
categ[col[ci]] = categs
ci+=1
df_train1.head()
C:\Users\zmakumbe\.conda\envs\wqu_ml_fin\lib\site-packages\pandas\core\indexing.py:670: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
iloc._setitem_with_indexer(indexer, value)
| MSSubClass | MSZoning | LotFrontage | LotArea | Street | LotShape | LandContour | Utilities | LotConfig | LandSlope | ... | EnclosedPorch | 3SsnPorch | ScreenPorch | PoolArea | MiscVal | MoSold | YrSold | SaleType | SaleCondition | SalePrice | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 60 | 3 | 65.0 | 8450 | 1 | 3 | 3 | 0 | 4 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 2 | 2008 | 8 | 4 | 208500 |
| 1 | 20 | 3 | 80.0 | 9600 | 1 | 3 | 3 | 0 | 2 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 5 | 2007 | 8 | 4 | 181500 |
| 2 | 60 | 3 | 68.0 | 11250 | 1 | 0 | 3 | 0 | 4 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 9 | 2008 | 8 | 4 | 223500 |
| 3 | 70 | 3 | 60.0 | 9550 | 1 | 0 | 3 | 0 | 0 | 0 | ... | 272 | 0 | 0 | 0 | 0 | 2 | 2006 | 8 | 0 | 140000 |
| 4 | 60 | 3 | 84.0 | 14260 | 1 | 0 | 3 | 0 | 2 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 12 | 2008 | 8 | 4 | 250000 |
5 rows × 76 columns
ci = 0
col = list(df_test1.columns)
categ = {}
for dt in df_test1.dtypes:
if dt == object:
categs= category_to_num(df_test1[col[ci]])
categ[col[ci]] = categs
ci+=1
plt.figure(figsize=(10,5))
y = df_train.SalePrice
sb.set_style('whitegrid')
plt.subplot(121)
sb.distplot(y)
df_train['SalePrice_log'] = np.log(df_train.SalePrice)
y2 = df_train.SalePrice_log
plt.subplot(122)
sb.distplot(y2)
plt.show()
C:\Users\zmakumbe\.conda\envs\wqu_ml_fin\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
C:\Users\zmakumbe\.conda\envs\wqu_ml_fin\lib\site-packages\seaborn\distributions.py:2557: FutureWarning: `distplot` is a deprecated function and will be removed in a future version. Please adapt your code to use either `displot` (a figure-level function with similar flexibility) or `histplot` (an axes-level function for histograms).
warnings.warn(msg, FutureWarning)
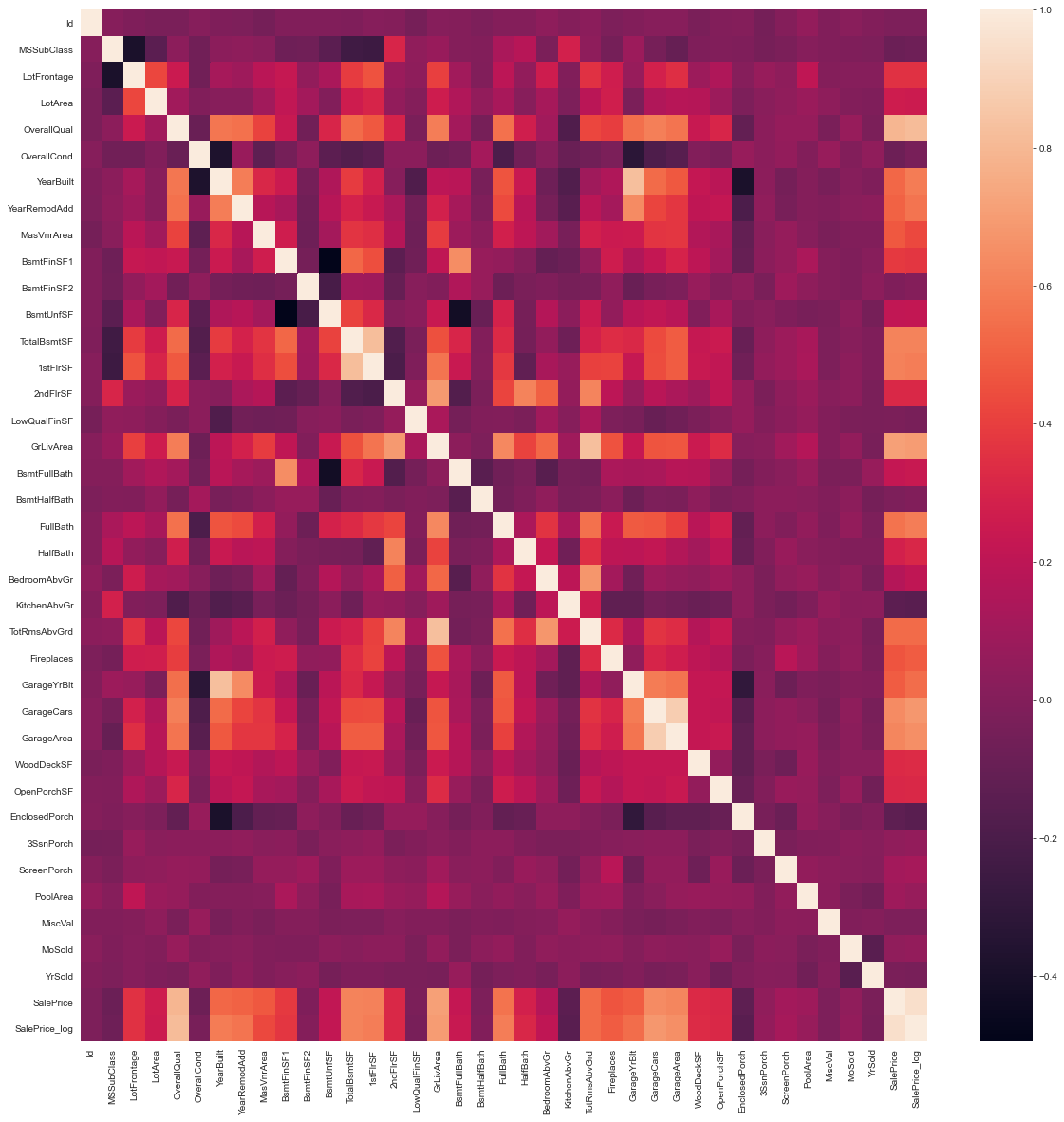
Data Correlation:
- It is important to determine any interdependencies if they exist.
# Lets explore the correlations in our data set
plt.figure(figsize=(20,20))
sb.heatmap(df_train.corr())
<AxesSubplot:>
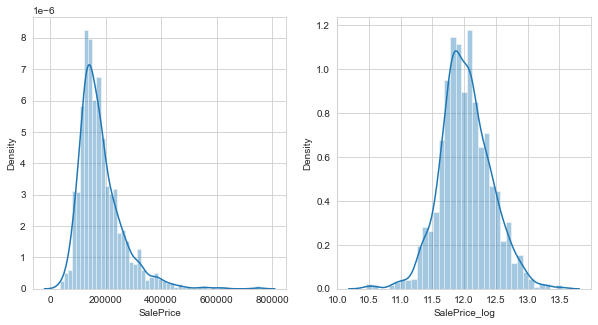
Visualising the output data
- Next, taking the Sale Price data (which will bo our output variable) we plot a bar graph
- From above, we find that the data is skewed but the log-transoformed data has a much better distribution
- Such transformations help us avoid having to remove outliers.
PCA Analysis of the data:
- Considering how many columns we have as well as the hunch we have pertaining to the ‘NaN’ values, we expect some columns to be redundant
- We conduct a Principal Component Analysis (PCA) in order to determine if a smaller set of the data can be used to determine the output
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
#Setting the input and output variables
x = df_train1[c[:-1]]
y = df_train1['SalePrice']
#Splitting the data into training and testing data for a trial run
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size = 0.2, random_state = 0)
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
# Applying PCA function on training
# and testing set of X component
pca = PCA(n_components = 50)
X_train = pca.fit_transform(X_train)
X_test = pca.transform(X_test)
explained_variance = pca.explained_variance_ratio_
print('%-age of variance explained by the 45 principal components')
np.round(explained_variance*100,1)
%-age of variance explained by the 45 principal components
array([13.7, 5.6, 4.9, 4. , 3. , 2.8, 2.4, 2.3, 2.2, 2.1, 2. ,
2. , 1.9, 1.8, 1.7, 1.6, 1.6, 1.6, 1.5, 1.5, 1.5, 1.5,
1.4, 1.4, 1.4, 1.3, 1.3, 1.2, 1.2, 1.2, 1.2, 1.1, 1.1,
1.1, 1. , 1. , 1. , 0.9, 0.9, 0.9, 0.9, 0.8, 0.8, 0.8,
0.8, 0.7, 0.7, 0.7, 0.7, 0.7])
plt.figure(figsize=(10,5))
plt.plot(np.cumsum(pca.explained_variance_ratio_) )
plt.xticks(np.arange(start=0, stop=len(pca.explained_variance_ratio_), step=1),rotation = 70)
plt.grid()
plt.show()
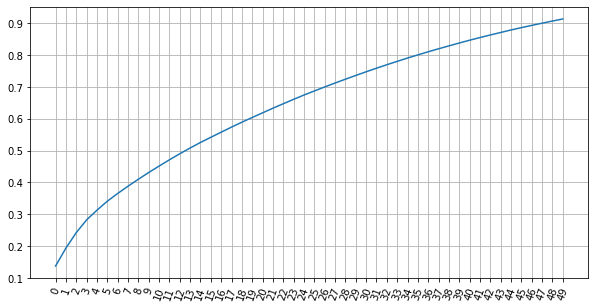
- After conducting the PCA analysis we find that we need at least
- 35 data columns to explain at least 80% of the variation in the data,
- 40 data columns to explain at least 85% of the variation in the data, and
- 47 data columns to explain at least 90% of the variation in the data
# Create linear regression object
regr = LinearRegression()
# Fit
regr.fit(X_train, y_train)
# Calibration
regr.score(X_test, y_test)
0.6565305727301065
pca = PCA(n_components = 50)
regr_pca = LinearRegression()
# Fit
X_pca_train = pca.fit_transform(X_train)
X_pca_test = pca.fit_transform(X_test)
regr_pca.fit(X_pca_train, y_train)
regr.score(X_pca_test, y_test)
#cross_val_score(regr_pca, X_pca_train, y_train).mean()
-1.0756078955375763
References:
- https://www.educative.io/edpresso/how-to-check-if-a-key-exists-in-a-python-dictionary
- https://towardsdatascience.com/predicting-house-prices-with-machine-learning-62d5bcd0d68f